Overview¶
A detailed description of the ideas behind the design of the toolbox can be found in our paper:
A mean-field toolbox for spiking neuronal network model analysis
NNMT can be used in two different ways: the basic workflow or the model workflow.
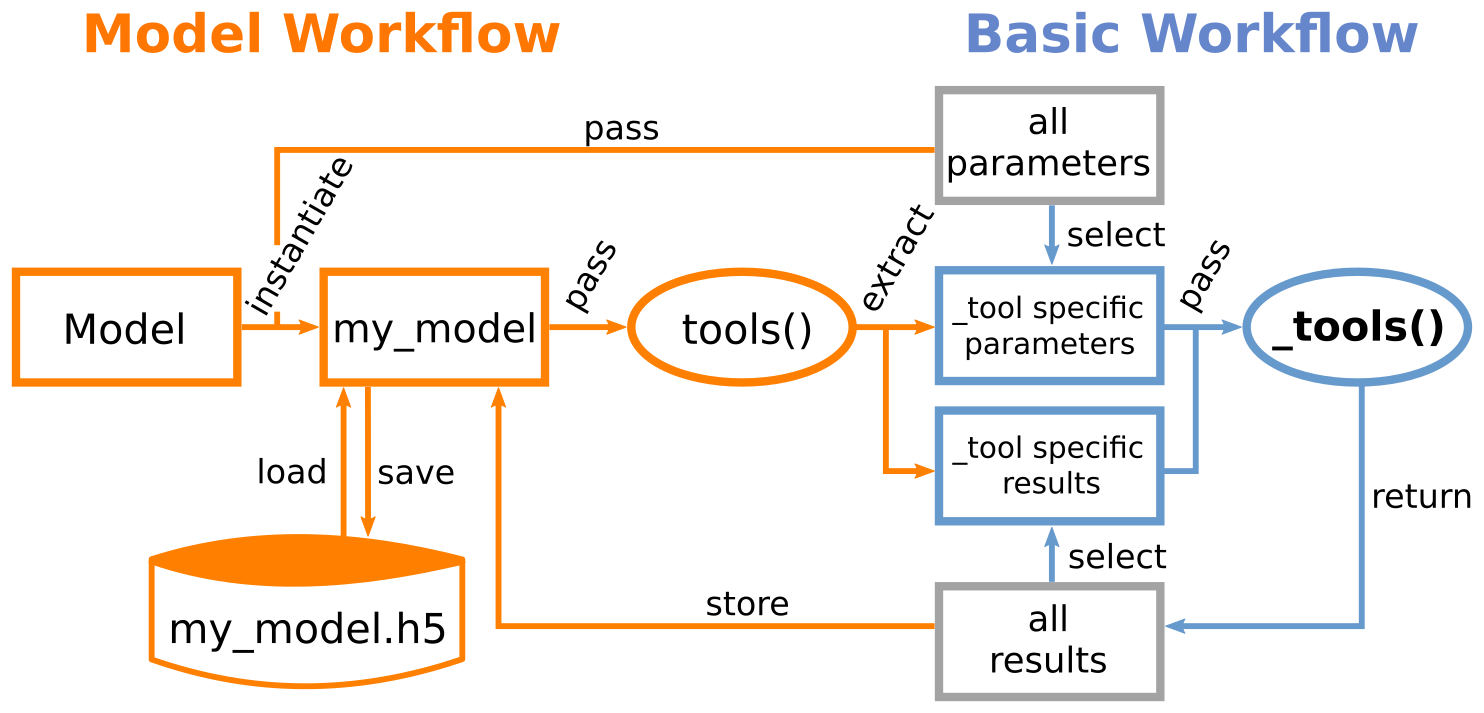
Basic workflow¶
The core of NNMT are the various implemented analytical methods for neuronal network model analysis. The simplest way to use those methods is to select one of the available Tools and use the corresponding underscored function or _tool. Those _tools take all required parameters directly as arguments for computing the desired results. This lightweight approach gives you full flexibility.
For example, calculating the firing rate for a random network of leaky
integrate-and-fire neurons with exponential synapses can be done using
nnmt.lif.exp._firing_rates():
rates = nnmt.lif.exp._firing_rates(J, K, V_0_rel, V_th_rel,
tau_m, tau_r, tau_s,
J_ext, K_ext, nu_ext)
However, certain _tools require a large number of inputs, which can make working with them a little cumbersome. To simplify their usage and support users unfamiliar with the underlying theory, we developed the more convenient model workflow.
Model workflow¶
Instead of passing the required arguments directly to the respective _tools, one can instantiate a Model and pass it to the corresponding non-underscored wrapper or tool.
Models basically are containers for network parameters, analysis parameters,
and results, with methods for changing parameters, saving, and loading results.
On instantiation they read in network and analysis parameters from dictionaries
or yaml files, calculate further depenend parameters from those, and
convert all parameters to SI units using the functionality of
Pint. Network parameters are all
parameters that describe properties of the network itself, like for example the
number of neurons in each population or the membrane time constants. Analysis
parameters are all parameters that do not describe properties of the network
but need to be defined in order to calculate quantities of interest. For
example, one needs to define the frequencies for which to calculate a frequency
dependent quantity.
When you call a tool, it tries to extract the parameters needed by the wrapped _tool. If you are missing some parameters for applying the tool you would like to use, you will receive an error message telling you which parameters you still need to define:
RuntimeError: You are missing 'tau_m' for calculating the firing rate!
Have a look into the documentation for more details on 'lif' parameters.
Sometimes, before you can calculate a quantity, you first have to calculate something else. If you did not perform the relevant computation previously, you will also receive an error message:
RuntimeError: You first need to calculate 'lif.exp.effective_connectivity'.
Once all arguments needed are available, the tools passes them to the wrapped
_tool, which calculates the result. Finally, the tool stores the computed
result in your models results dictionary and returns the result. If you try
to compute the same quantity repeatedly with identical parameters, a tool will
detect this and return the cached result instead.
Returning to the firing rate example from above, we can conduct the same
computation with nnmt.lif.exp.firing_rates() and a
nnmt.models.Basic model:
network = nnmt.models.Basic('network_params.yaml')
rates = nnmt.lif.exp.firing_rates(network)
where we defined the respective parameters in yaml files in the following
format:
# number of neurons in each population
N:
- 1000
- 2000
# membrane tim constant
tau_m:
val: 10
unit: ms
# external input
nu_ext:
val:
- 1
- 2
unit: Hz
...
Package structure¶
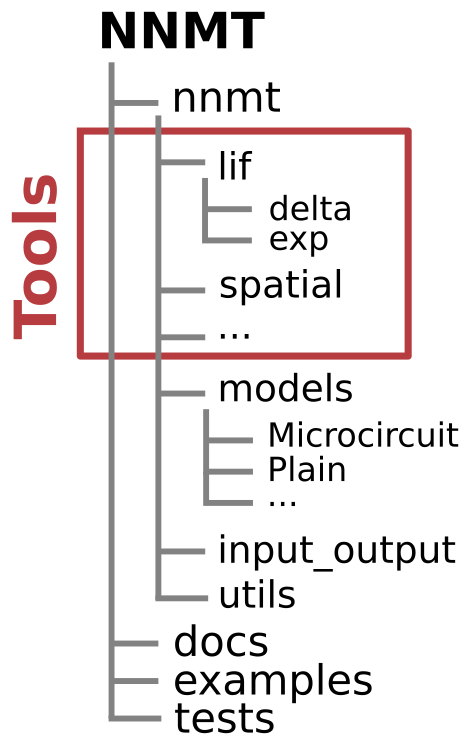
In the submodule nnmt.models you can find all models as well as the generic
nnmt.models.Network class, which all models are derived from.
nnmt.input_output contains helper routines for input and output related
tasks, including a basic wrapper of the h5py package, which allows you to
store and load dictionaries in h5 files.
nnmt.utils contains utility routines, primarily for tool developers,
although some of the functions may be useful for regular users as well, such as
nnmt.utils._convert_to_si_and_strip_units().
All other submodules, like nnmt.lif and nnmt.network_properties
contain the tools.
Note that we have a bunch of nice examples, as well as an extensive test suite, and a contributors’ guide with further information on the toolbox.